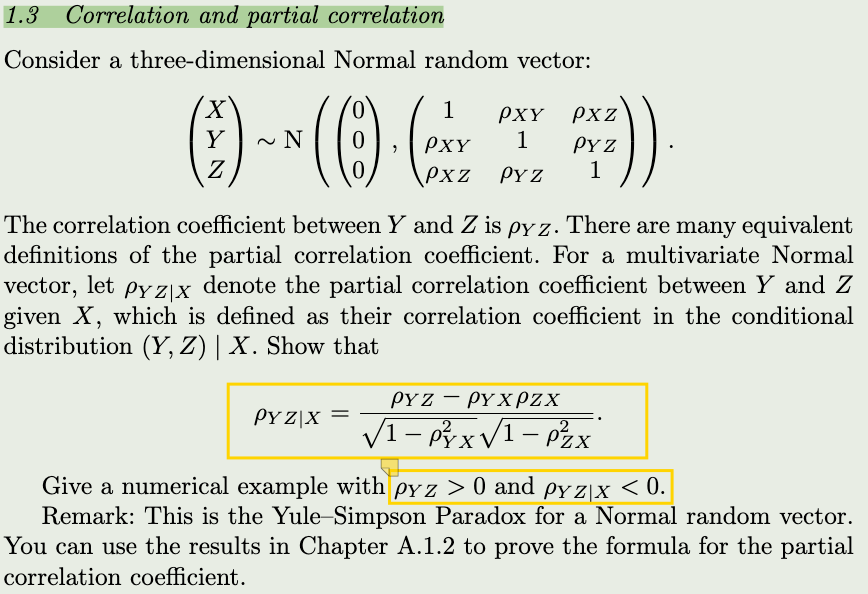
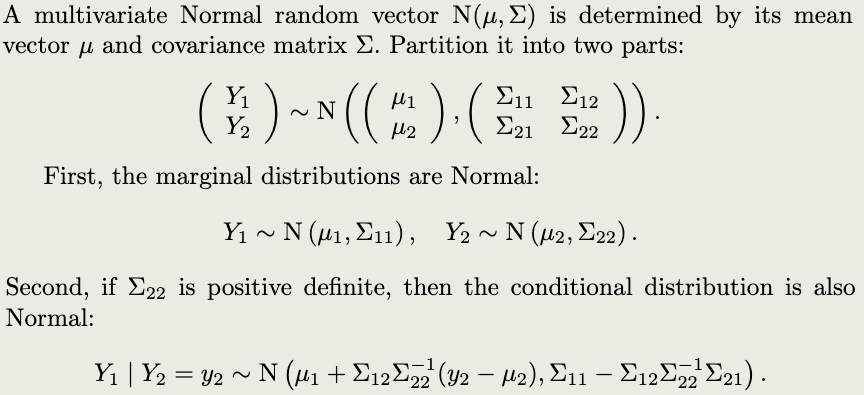
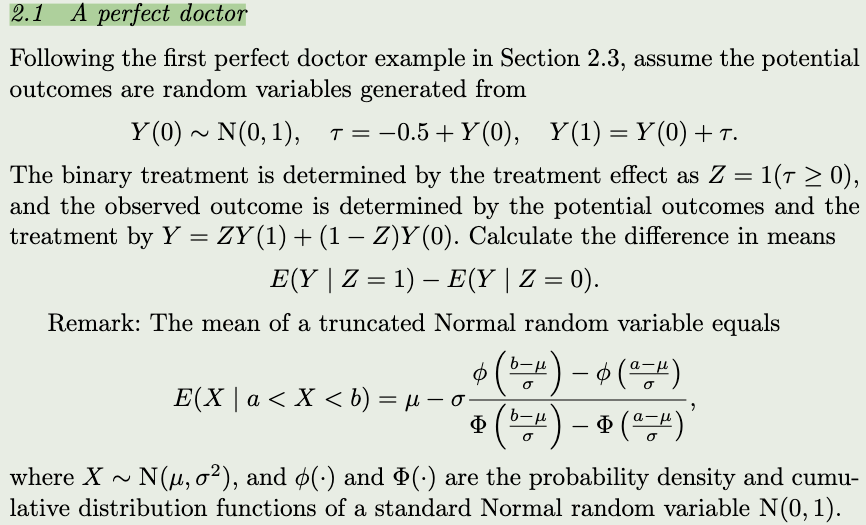
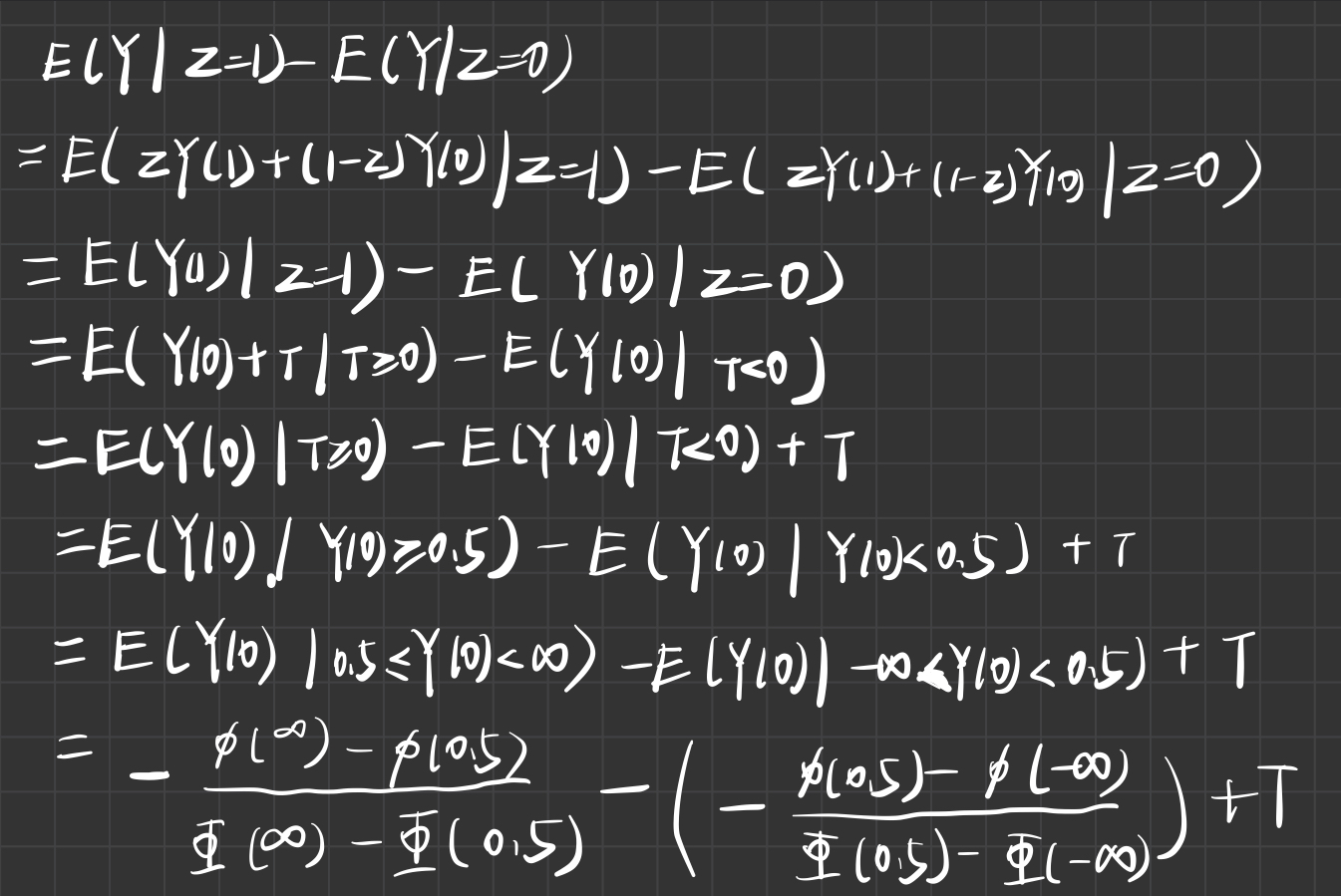z <- rbinom(10000,1,0.5)
y <- rbinom(10000,1,0.3)
t <- table(z,y)
rd <- function(x){x[1,1]/(x[1,2]+x[1,1])-x[2,1]/(x[2,2]+x[2,1])}
rr <- function(x){(x[1,1]/(x[1,2]+x[1,1]))/(x[2,1]/(x[2,2]+x[2,1]))}
or <- function(x){(x[1,1]*x[2,2])/(x[2,1]*x[1,2]) }
rd(t)
z <- rbinom(10000,1,0.5)
y <- rbinom(10000,1,z*0.5+0.1)
t <- table(z,y)
rd <- function(x){x[1,1]/(x[1,2]+x[1,1])-x[2,1]/(x[2,2]+x[2,1])}
rr <- function(x){(x[1,1]/(x[1,2]+x[1,1]))/(x[2,1]/(x[2,2]+x[2,1]))}
or <- function(x){(x[1,1]*x[2,2])/(x[2,1]*x[1,2]) }
rd(t)